Clean Corpora and Datasets are a Major Part of the Effort

With our discussions of network analysis and knowledge extractions from our knowledge graph now behind us, we are ready to tackle the questions of analytic applications and machine learning in earnest for our Cooking with Python and KBpedia series. We will be devoting our next nine installments to this area. We devote two installments to data sources and input preparations, largely based on NLP (natural language processing) applications. Then we devote two installments to ‘standard’ machine learning (largely) using the scikit-learn packages. We next devote four installments to deep learning, split equally between the Deep Learning Graph (DGL) and PyTorch Geometric (PyG) frameworks. We conclude this Part VI with a summary and comparison of results across these installments based on the task of node classification.
In this particular installment we flesh out the plan for completing these installments and discuss data sources and completing data prep needed for the plan. We provide particular attention to the architecture and data flows within the PyTorch framework. We describe the additional Python packages we need for this work, and install and configure the first ones. We discuss general sources of data and corpora useful for machine learning purposes. Our coding efforts in this installment will obtain and clean the Wikipedia pages that supplement the two structural and annotation sources based on KBpedia that were covered in the prior installment. These three sources of structure, annotations and pages are the input basis to creating our own embeddings to be used in many of the machine learning tests.
Plan for Completion of Part VI
The broad ecosystem of Python packages I was considering looked, generally, to be good choices to work together, as first outlined in CWPK #61. I had done an adequate initial diligence. But, how all of this was to unfold, what my plan of attack should be, became driving factors I had to solve to shorten my development and coding efforts. So, with an understanding of how we could extract general information from KBpedia useful to analysis and machine learning, I needed to project out over the entire anticipated scope to see if, indeed, these initial sources looked to be the right ones for our purposes. And, if so, how shall the efforts be sequenced and what is the flow of data?
Much reading and research went into this effort. It is true, for example, that we had already prepared a pretty robust series of analytic and machine learning case studies in Clojure, available from the KBpedia Web site. I revisited each of these use cases and got some ideas of what made sense for us to attempt with Python. But I needed to understand the capabilities now available to us with Python, so I also studied each of the candidate keystone packages in some detail.
I will weave the results of this research as the next installments unfold, providing background discussion in context and as appropriate. But, in total, I formulated about 30 tasks going forward that appeared necessary to cover the defined scope. The listing below summarizes these steps, and keys the transition point (as indicated by CWPK installment number) for proceeding to each next new installment:
- Formulate Part VI plan
- Extract two source files from KBpedia
- structure
- annotations
- Set environment up (not doing virtual)
- Obtain Wikipedia articles for matching RCs
- Set up gensim
- Clean Wikipedia articles, all KB annotations
- Set up spaCy
- ID, extract phrases
- Finish embeddings prep #64
- remove stoplist
- create numeric??
- Create embedding models:
- word2vec and doc2vec
- Text summarization for short articles (gensim)
- Named entity recognition
- Set up scikit-learn #65
- Create master pandas file
- Do event/action extraction
- Do scikit-learn classifier #66
- SVM
- k-nearest neighbors
- random forests
- Introduce the sklearn.metrics module and confusion matrix, etc. The standard for reporting
- Discuss basic test parameters/’gold standars’
- Knowledge graph embeddings #67
- Create embedding models -2
- KB-struct
- KB-annot
- KB-annot-full: what is above + below
- KB-annot-page
- Set up PyTorch/DLG-KE #68
- Set up PyTorch/PyG
- Formulate learning pathway/code
- Do standard DL classifiers: #69
- TransE
- TransR
- RESCAL
- DistMult
- ComplEx
- RotatE
- Do research DL classifiers: #70
- VAE
- GGSNN
- MPNN
- ChebyNet
- GCN
- SAGE
- GAT
- Choose a model evaluator: #71
- scikit-learn
- pyTorch
- other?
- Collate prior results
- Evaluate prior results
- Present comparative results
Some of these steps also needed some preliminary research before proceeding. For example, knowing I wanted to compare results across algorithms meant I needed to have a good understanding of testing and analysis requirements before starting any of the tests.
PyTorch Architecture
A critical question in contemplating this plan was how exactly data needed to be produced, staged, and then fed into the analysis portions. From the earlier investigations I had identified the three categories of knowledge grounded in KBpedia that could act as bases or features to machine learning; namely, structure, annotations and pages. I also had identified PyTorch as a shared abstraction layer for deep and machine learning.
I was particularly focused on the question of data formats and representations such that information could be readily passed from one step to the next in the analysis pipeline. Figure 1 is the resulting data flow chart and architecture that arose from these investigations.
First, the green block labeled ‘owlready2’ represents that Python package, but also the location where the intact knowledge graph of KBpedia is stored and accessed. As early installments covered, we can use either owlready2 or Protégé to manage this knowledge graph, though owlready2 is the point at which the KBpedia information is exported or extracted for downstream uses, importantly machine learning. As our owlready2 discussions also indicated, there is a close relationship between it and RDFLib (which is also the SPARQL access point). RDFLib can provide direct input into NetworkX, but that is limited to structure only.
The clearest common denominator format for entry into the machine learning pipeline is pandas via CSV files. This centrality is fortunate given that all of our prior KBpedia extract-and-build routines have been designed around this format. This format is also one of the direct feeds possible into the PyTorch datasets format, as the figure shows:
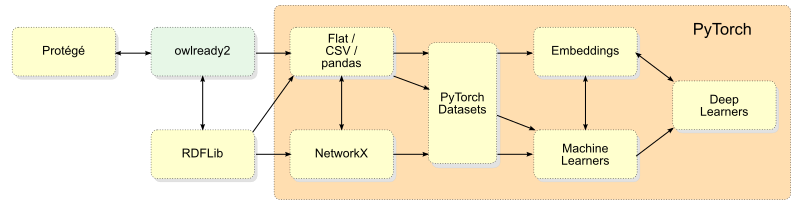
An important block on the figure is for ’embeddings’. If you recall, all text needs to first be encoded to a numeric form to be understood by the computer. This process can also undertake dimensionality reduction, important for a sparse matrix data form like language. This same ability can be applied to graph structure and interactions. Thus, the ’embedding’ block is a pivotal point at which we can represent words, sentences, paragraphs, documents, nodes, or entire graphs. We will focus much on embeddings throughout this Part VI.
For training purposes we can also feed pre-trained corpora or embeddings into the system. We address this topic in the next main section.
Figure 1 is not meant to be a comprehensive view of PyTorch, but it is one useful to understand data flows with respect to our use of the KBpedia knowledge graph. Over the course of this research, I also encountered many PyTorch-related extensions that, when warranted, I include in the discussion.
Possible Extensions
There are some extensions to the PyTorch ecosystem that we will not be using or testing in this CWPK series. Here are some of the ones that seem closest in capabilities to what we are doing with KBpedia:
- PyCaret is essentially a Python wrapper around several machine learning libraries and frameworks such as scikit-learn, XGBoost, Microsoft LightGBM, spaCy, and many more
- PiePline is a neural networks training pipeline based on PyTorch. Designed to standardize training process and accelerate experiments
- Catalyst helps to write full-featured deep learning pipelines in a few lines of code
- Poutyne is a Keras-like framework for PyTorch and handles much of the boilerplating code needed to train neural networks
- torchtext has some capabilities in language modeling, sentiment analysis, text classification, question classification, entailment, machine translation, sequence tagging, question answering, and unsupervised learning
- Spotlight uses PyTorch to build both deep and shallow recommender models.
Corpora and Datasets
There are many off-the-shelf resources that can be of use when doing machine learning involving text and language. (There are as well for images, but that is out of scope to our current interests.) These resources fall into three main areas:
- corpora – are language resources of either a general or domain nature, with vetted relationships or annotations between terms and concepts or other pre-processing useful to computational linguistics
- pre-trained models – are pre-calculated language models, often expressing probability distributions over words or text. Some embeddings can act in this manner. Transformers use deep learning to train their representations, with BERT being a notable example
- embeddings – are vector representations of chunks of text, ranging from individual words up to entire documents or languages. The numeric representation either represents a pooled statistical representation across all tokens (the so-called CBOW approach) or context and adjacency using the skip-gram or similar method. GloVe, word2vec and fastText are example methodologies for producing word embeddings.
Example corpora include Wikipedia (in multiple languages), news articles, Web crawls, and many others. Such corpora can be used as the language input basis for training various models, or may be a reference vocabulary for scoring and ranking input text. Various pre-trained language models are available, and embedding methods are available in a number of Python packages, including scikit-learn, gensim and spaCy used in cowpoke.
Pre-trained Resources
There are a number of free or open-source resources for these corpora or datasets. Some include:
- Transformers provides thousands of pretrained models to perform tasks on texts such as classification, information extraction, question answering, summarization, translation, text generation, etc in 100+ languages
- HuggingFace datasets
- English Wikipedia dump
- Wikipedia2Vec pre-trained embeddings
- gensim datasets contain links to 8 options
- word2vec pre-trained models lists 16 or so datasets
- A comprehensive list of available gensim datasets and models
- 11 pre-trained word embedding models in various embedding formats
- Some older GloVe embeddings
- Word vectors for 157 languages
- DBpedia entity typing + word embeddings.
Setting Up the Environment
In doing this research, I also assembled the list of Python packages needed to add these capabilities to cowpoke. Had I not just updated the conda packages, I would do so now:
conda update --all
Next, the general recommendation when installing multiple new packages in Python is to do them in one batch, which allows the package manager (conda in our circumstance) to check on version conflicts and compatibility during the install process. However, with some of the packages involved in the current expansion, there are other settings necessary that obviates this standard ‘batch’ install recommendation.
Another note is important here. In an enterprise environment with many Python projects, it is also best to install these machine learning extensions into their own virtual environment. (I covered this topic a bit in CWPK #58.) However, since we are keeping this entire series in its own environment, we will skip that step here. You may prefer the virtual option.
So, we will begin with those Python packages and frameworks that pose their own unique set-up and install challenges. We begin with PyTorch. We need to first appreciate that the rationale for PyTorch was to abstract machine learning constructs while taking advantage of graphics processing units (GPUs) (specifically, Nvidia via the CUDA interface). The CUDA architecture provides one or two orders of magnitude speed up on a local machine. Unfortunately, my local Windows machine does not have the separate Nvidia GPU, so I want to install the no CUDA option. For the PyTorch install options, visit https://pytorch.org/get-started/locally/. This figure shows my selections prior to download (yours may vary):
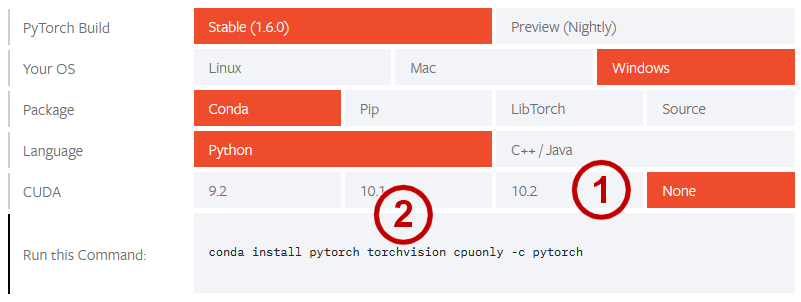
In my circumstance, my local machine does not have a separate graphics processor, so I set the CUDA requirement to ‘None’ (1). I also removed the ‘torchvision’ command line specification (2) since that is an image-related package. (We may later need some libraries from this package, in which case we will then install it.) The PyTorch package is rather large, so install takes a few minutes. Here is the actual install command:
conda install pytorch cpuonly -c pytorch
Since we were not able to batch all new packages, I decide to continue with some of the other major additions in a sequential matter, with spaCy and its installation next:
conda install -c conda-forge spacy
and then gensim and its installation:
conda install -c conda-forge gensim
and then DLG, which has an installation screen similar to PyTorch in Figure 2 with the same picked options:
conda install -c dglteam dgl
The DLG-KE extension needs to be built from source for Windows, so we will hold off on that now until we need it. We next install PyTorch Geometric, which needs to be installed from a series of binaries, with CPU or GPU individually specified:
pip install torch-scatter==latest+cpu -f https://pytorch-geometric.com/whl/torch-1.6.0.html
pip install torch-sparse==latest+cpu -f https://pytorch-geometric.com/whl/torch-1.6.0.html
pip install torch-cluster==latest+cpu -f https://pytorch-geometric.com/whl/torch-1.6.0.html
pip install torch-spline-conv==latest+cpu -f https://pytorch-geometric.com/whl/torch-1.6.0.html
pip install torch-geometric
These new packages join these that are already a part of my local conda packages, and which will arise in the coming installments:
scikit-learn and tqdm.
Getting Wikipedia Pages
With these preliminaries complete, we are now ready to resume our data preparation tasks for our embedding and machine learning experiments. In the prior installment, we discussed two of the three source files we had identified for these efforts, the KBpedia structure (kbpedia/v300/extractions/data/graph_specs.csv) and the KBpedia annotations (kbpedia/v300/extractions/classes/Generals_annot_out.csv) files. In this specific section we obtain the third source file of pages from Wikipedia.
Of the 58,000 reference concepts presently contained in KBpedia, about 45,000 have a directly corresponding Wikipedia article or listing of category articles. These provide a potentially rich source of content for language models and embeddings. The challenge is how to obtain this content in a way that can be readily processed for our purposes.
We have been working with Wikipedia since its inception, so we knew that there are data sources for downloads or dumps. For example, the periodic language dumps such as https://dumps.wikimedia.org/enwiki/20200920/ may be accessed to obtain full-text versions of articles. Such dumps have been used scores of times to produce Wikipedia corpora in many different languages and for many different purposes. But, our own mappings are a mere subset, about 1% of the nearly 6 million articles in the English Wikipedia alone. So, even if we grabbed the current dump or one of the corpora so derived, we would need to process much content to obtain the subset of interest.
Unfortunately, Wikipedia does not have a direct query or SPARQL form as exists for Wikidata (which also does not have full-text articles). We could obtain the so-called ‘long abstracts’ of Wikipedia pages from DBpedia (see, for example, https://wiki.dbpedia.org/downloads-2016-10), but this source is dated and each abstract is limited to about 220 words; further, a full download of the specific file in English is about 15 GB!
The basic approach, then, appeared that I would need to download the full Wikipedia article file, figure out how to split it into parts, and then match identifiers between KBpedia mappings and the full dataset to obtain the articles of interest. This approach is not technically difficult, but it is a real pain in the ass.
So, shortly before I committed to this work effort, I challenged myself to find another way that was perhaps less onerous. Fortunately, I found the online Wikipedia service, https://en.wikipedia.org/wiki/Special:Export, that allows one to submit article names to a text box and then get the full page article back in XML format. I tested this online service with a few articles, then 100, and then ramped up to a listing of 5 K at a time. (Similar services often have governors that limit the frequency or amounts of individual requests.) This approach worked!, and within 30 min I had full articles in nine separate batches for all 45 K items in KBpedia.
Clean All Input Text
This file is a single article from the Wikipedia English dump for 1-(2-Nitrophenoxy)octane:
<page>
<title>1-(2-Nitrophenoxy)octane</title>
<ns>0</ns>
<id>11793192</id>
<revision>
<id>891140188</id>
<parentid>802024542</parentid>
<timestamp>2019-04-05T23:04:47Z</timestamp>
<contributor>
<username>Koavf</username>
<id>205121</id>
</contributor>
<minor/>
<comment>/* top */Replace HTML with MediaWiki markup or templates, replaced: <sub> → {{sub| (3), </sub> → }} (3)</comment>
<model>wikitext</model>
<format>text/x-wiki</format>
<text bytes="2029" xml:space="preserve">{{chembox
| Watchedfields = changed
| verifiedrevid = 477206849
| ImageFile =Nitrophenoxyoctane.png
| ImageSize =240px
| ImageFile1 = 1-(2-Nitrophenoxy)octane-3D-spacefill.png
| ImageSize1 = 220
| ImageAlt1 = NPOE molecule
| PIN = 1-Nitro-2-(octyloxy)benzene
| OtherNames = 1-(2-Nitrophenoxy)octane<br />2-Nitrophenyl octyl ether<br />1-Nitro-2-octoxy-benzene<br />2-(Octyloxy)nitrobenzene<br />Octyl o-nitrophenyl ether
|Section1={{Chembox Identifiers
| Abbreviations =NPOE
| ChemSpiderID_Ref = {{chemspidercite|correct|chemspider}}
| ChemSpiderID = 148623
| InChI = 1/C14H21NO3/c1-2-3-4-5-6-9-12-18-14-11-8-7-10-13(14)15(16)17/h7-8,10-11H,2-6,9,12H2,1H3
| InChIKey = CXVOIIMJZFREMM-UHFFFAOYAD
| StdInChI_Ref = {{stdinchicite|correct|chemspider}}
| StdInChI = 1S/C14H21NO3/c1-2-3-4-5-6-9-12-18-14-11-8-7-10-13(14)15(16)17/h7-8,10-11H,2-6,9,12H2,1H3
| StdInChIKey_Ref = {{stdinchicite|correct|chemspider}}
| StdInChIKey = CXVOIIMJZFREMM-UHFFFAOYSA-N
| CASNo_Ref = {{cascite|correct|CAS}}
| CASNo =37682-29-4
| PubChem =169952
| SMILES = [O-][N+](=O)c1ccccc1OCCCCCCCC
}}
|Section2={{Chembox Properties
| Formula =C{{sub|14}}H{{sub|21}}NO{{sub|3}}
| MolarMass =251.321
| Appearance =
| Density =1.04 g/mL
| MeltingPt =
| BoilingPtC = 197 to 198
| BoilingPt_notes = (11 mm Hg)
| Solubility =
}}
|Section3={{Chembox Hazards
| MainHazards =
| FlashPt =
| AutoignitionPt =
}}
}}
'''1-(2-Nitrophenoxy)octane''', also known as '''nitrophenyl octyl ether''' and abbreviated '''NPOE''', is a
[[chemical compound]] that is used as a matrix in [[fast atom bombardment]] [[mass spectrometry]], liquid
[[secondary ion mass spectrometry]], and as a highly [[lipophilic]] [[plasticizer]] in [[polymer]]
[[Polymeric membrane|membranes]] used in [[ion selective electrode]]s.
== See also ==
* [[Glycerol]]
* [[3-Mercaptopropane-1,2-diol]]
* [[3-Nitrobenzyl alcohol]]
* [[18-Crown-6]]
* [[Sulfolane]]
* [[Diethanolamine]]
* [[Triethanolamine]]
{{DEFAULTSORT:Nitrophenoxy)octane, 1-(2-}}
[[Category:Nitrobenzenes]]
[[Category:Phenol ethers]]</text>
<sha1>0n15t2w0sp7a50fjptoytuyus0vsrww</sha1>
</revision>
</page>We want to extract out the specific article text (denoted by the <text> field), perhaps capture some other specific fields, remove internal tags, and then create a clean text representation that we can further process. This additional processing includes removing stoplist words, finding and identifying phrases (multiple token chunks), and then tokenizing the text suitable for processing as computer input.
There are multiple methods available for this kind of processing. One approach, for example, uses XML parsing and specific code geared to the Wikipedia dump. Another approach uses a dedicated Wikipedia extractor. There are actually a few variants of dedicated extractors.
However, one particular Python package, gensim, provides multiple utilities and Wikipedia services. Since I had already identified gensim to provide services like sentiment analysis and some other NLP capabilities, I chose to focus on using this package for the needed Wikipedia cleaning tasks.
Gensim has a gensim.corpora.wikicorpus.WikiCorpus class designed specifically for processing the Wikipedia article dump file. Fortunately, I was able to find some example code on KDnuggets that showed the way in how to process this file
- https://stackoverflow.com/questions/56715394/how-do-i-use-the-wikipedia-dump-as-a-gensim-model (doc2vec example) and
- https://www.kdnuggets.com/2017/11/building-wikipedia-text-corpus-nlp.html as another.
However, prior to using gensim, I needed to combine the batch outputs from my Wikipedia page retrievals into a single xml file, which I could then bzip for direct ingest by gensim. (Most gensim models and capabilities can read either bzip or text files.)
Each 5 K xml page retrieval from Wikipedia comes with its own header and closing tags. These need to be manually snipped out of the group retrieval files before combining. We prepared these into nine blocks that corresponded to each of the batch Wikipedia retrievals, and retained the header and closing tags in the first and last files respectively:
out_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-pages-full.xml'
filenames = [r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-1.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-2.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-3.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-4.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-5.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-6.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-7.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-8.txt',
r'C:\1-PythonProjects\kbpedia\v300\models\inputs\Wikipedia-pages-9.txt']
with open(out_f, 'w', encoding='utf-8') as outfile:
for fname in filenames:
with open(fname, encoding='utf-8', errors='ignore') as infile:
i = 0
for line in infile:
i = i + 1
try:
outfile.write(line)
except Exception as e:
print('Error at line:' + i + str(e))
print('Now combined:' + fname)
outfile.close
print('Now all files combined!') The output of this routine is then bzipped offline, and then used as the submission to the gensim WikiCorpus function that processes the standard xml output:
"""
Creates a corpus from Wikipedia dump file.
Inspired by:
https://github.com/panyang/Wikipedia_Word2vec/blob/master/v1/process_wiki.py
"""
import sys
from gensim.corpora import WikiCorpus
in_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-pages-full.xml.bz2'
out_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-output-full.txt'
def make_corpus(in_f, out_f):
"""Convert Wikipedia xml dump file to text corpus"""
output = open(out_f, 'w', encoding='utf-8') # made change
wiki = WikiCorpus(in_f)
i = 0
for text in wiki.get_texts():
try:
output.write(' '.join(map(lambda x:x.decode('utf-8'), text)) + '\n')
except Exception as e:
print ('Exception error: ' + str(e))
i = i + 1
if (i % 10000 == 0):
print('Processed ' + str(i) + ' articles')
output.close()
print('Processed ' + str(i) + ' articles;')
print('Processing complete!')
make_corpus(in_f, out_f)We further make a smaller input file, enwiki-test-corpus.xml.bz2, with only a few records from the Wikipedia XML dump in order to speed testing of the above code.
Initial Results
Here is what the sample program produced for the entry for 1-(2-Nitrophenoxy)octane listed above:
nitrophenoxy octane also known as nitrophenyl octyl ether and
abbreviated npoe is chemical compound that is used as matrix in fast
atom bombardment mass spectrometry liquid secondary ion mass
spectrometry and as highly lipophilic plasticizer in polymer membranes
used in ion selective electrodes see also glycerol mercaptopropane diol
nitrobenzyl alcohol crown sulfolane diethanolamine triethanolamine
We see a couple of things that are perhaps not in keeping with the extracted information we desire:
- No title
- No sentence boundaries
- No internal category links
- No infobox specifications
On the other hand, we do get the content from the ‘See Also’ section.
We want sentence boundaries for cleaner training purposes for word embedding models like word2vec. We want the other items so as to improve the lexical richness and context for the given concept. Further, we want two versions: one with titles as a separate field and one for learning purposes that includes the title in the lexicon (titles, after all, are preferred labels and deserve an additional frequency boost).
OK, so how does one make these modifications? My first hope was that arguments to these functions (args) might provide the specification latitude to deal with these changes. Unfortunately, none of the specified items fell into this category, though there is much latitude to modify underlying procedures. The second option was to find some third-party modification or override. Indeed, I did find one, that I found quite intriguing as a way to at least deal with sentence boundaries and possibly other areas. I spent nearly a full day trying to adapt this script, never succeeding. One fix would lead to another need for a fix, research on that problem, and then a fix and more problems. I’m sure most all of this is due to my amateur programming skills.
Still, the effort was frustrating. The good thing, however, is that in trying to work out a third-party fix, I was learning the underlying module. Eventually, it became clear if I was to address all desired areas it was smartest to modify the source directly. The three key functions that emerged as needing attention were tokenize, process_article and the class WikiCorpus(TextCorpus) code. In fact, it was the text processing heart of the last class that was the focus for changes, but the other two functions got involved because of their supporting roles. As I attempted to sub-class this basis with my own parallel approach (class KBWikiCorpus(WikiCorpus), I kept finding the need to bring into the picture more supporting functions. Some of this may have been due to nuances in how to specify imported functions and modules, which I am still learning about (see concluding installments). But it is also difficult to sub-set or modify any code.
The real impact of these investigations was to help me understand the underlying module. What at first blush looked too intimidating, now was becoming understandable. I could also see other portions of the underlying module that addressed ALL aspects of my earlier desires. Third-party modifications choose their own scope; direct modification of the underlying module provides more aspects to tweak. So, I switched emphasis from modifying a third-party overlay to directly changing the core underlying module.
Modifying WikiCorpus
We already knew the key functions needing focus. All changes to be made occur in the wikicorpus.py file that resides in your gensim package directory under Python packages. So, I make a copy of the original and name it such, then proceed to modify the base file. Though we will substitute this modified wikicorpus_kb.py file, I will also keep a backup of it as well such that we have copies of the original and modified file.
Here is the resulting modified code, with notes about key changes following the listing:
with open('files/wikicorpus_kb.py', 'r') as f:
print(f.read())#!/usr/bin/env python
# -*- coding: utf-8 -*-
#
# Copyright (C) 2010 Radim Rehurek <radimrehurek@seznam.cz>
# Copyright (C) 2012 Lars Buitinck <larsmans@gmail.com>
# Copyright (C) 2018 Emmanouil Stergiadis <em.stergiadis@gmail.com>
# Licensed under the GNU LGPL v2.1 - http://www.gnu.org/licenses/lgpl.html
"""Construct a corpus from a Wikipedia (or other MediaWiki-based) database dump.
Uses multiprocessing internally to parallelize the work and process the dump more quickly.
Notes
-----
If you have the `pattern <https://github.com/clips/pattern>`_ package installed,
this module will use a fancy lemmatization to get a lemma of each token (instead of plain alphabetic tokenizer).
See :mod:`gensim.scripts.make_wiki` for a canned (example) command-line script based on this module.
"""
import bz2
import logging
import multiprocessing
import re
import signal
from pickle import PicklingError
# LXML isn't faster, so let's go with the built-in solution
try:
from xml.etree.cElementTree import iterparse
except ImportError:
from xml.etree.ElementTree import iterparse
from gensim import utils
# cannot import whole gensim.corpora, because that imports wikicorpus...
from gensim.corpora.dictionary import Dictionary
from gensim.corpora.textcorpus import TextCorpus
from six import raise_from
logger = logging.getLogger(__name__)
ARTICLE_MIN_WORDS = 50
"""Ignore shorter articles (after full preprocessing)."""
# default thresholds for lengths of individual tokens
TOKEN_MIN_LEN = 2
TOKEN_MAX_LEN = 15
RE_P0 = re.compile(r'<!--.*?-->', re.DOTALL | re.UNICODE)
"""Comments."""
RE_P1 = re.compile(r'<ref([> ].*?)(</ref>|/>)', re.DOTALL | re.UNICODE)
"""Footnotes."""
RE_P2 = re.compile(r'(\n\[\[[a-z][a-z][\w-]*:[^:\]]+\]\])+$', re.UNICODE)
"""Links to languages."""
RE_P3 = re.compile(r'{{([^}{]*)}}', re.DOTALL | re.UNICODE)
"""Template."""
RE_P4 = re.compile(r'{{([^}]*)}}', re.DOTALL | re.UNICODE)
"""Template."""
RE_P5 = re.compile(r'\[(\w+):\/\/(.*?)(( (.*?))|())\]', re.UNICODE)
"""Remove URL, keep description."""
RE_P6 = re.compile(r'\[([^][]*)\|([^][]*)\]', re.DOTALL | re.UNICODE)
"""Simplify links, keep description."""
RE_P7 = re.compile(r'\n\[\[[iI]mage(.*?)(\|.*?)*\|(.*?)\]\]', re.UNICODE)
"""Keep description of images."""
RE_P8 = re.compile(r'\n\[\[[fF]ile(.*?)(\|.*?)*\|(.*?)\]\]', re.UNICODE)
"""Keep description of files."""
RE_P9 = re.compile(r'<nowiki([> ].*?)(</nowiki>|/>)', re.DOTALL | re.UNICODE)
"""External links."""
RE_P10 = re.compile(r'<math([> ].*?)(</math>|/>)', re.DOTALL | re.UNICODE)
"""Math content."""
RE_P11 = re.compile(r'<(.*?)>', re.DOTALL | re.UNICODE)
"""All other tags."""
RE_P12 = re.compile(r'(({\|)|(\|-(?!\d))|(\|}))(.*?)(?=\n)', re.UNICODE)
"""Table formatting."""
RE_P13 = re.compile(r'(?<=(\n[ ])|(\n\n)|([ ]{2})|(.\n)|(.\t))(\||\!)([^[\]\n]*?\|)*', re.UNICODE)
"""Table cell formatting."""
RE_P14 = re.compile(r'\[\[Category:[^][]*\]\]', re.UNICODE)
"""Categories."""
RE_P15 = re.compile(r'\[\[([fF]ile:|[iI]mage)[^]]*(\]\])', re.UNICODE)
"""Remove File and Image templates."""
RE_P16 = re.compile(r'\[{2}(.*?)\]{2}', re.UNICODE)
"""Capture interlinks text and article linked"""
RE_P17 = re.compile(
r'(\n.{0,4}((bgcolor)|(\d{0,1}[ ]?colspan)|(rowspan)|(style=)|(class=)|(align=)|(scope=))(.*))|'
r'(^.{0,2}((bgcolor)|(\d{0,1}[ ]?colspan)|(rowspan)|(style=)|(class=)|(align=))(.*))',
re.UNICODE
)
"""Table markup"""
IGNORED_NAMESPACES = [
'Wikipedia', 'Category', 'File', 'Portal', 'Template',
'MediaWiki', 'User', 'Help', 'Book', 'Draft', 'WikiProject',
'Special', 'Talk'
]
"""MediaWiki namespaces that ought to be ignored."""
def filter_example(elem, text, *args, **kwargs):
"""Example function for filtering arbitrary documents from wikipedia dump.
The custom filter function is called _before_ tokenisation and should work on
the raw text and/or XML element information.
The filter function gets the entire context of the XML element passed into it,
but you can of course choose not the use some or all parts of the context. Please
refer to :func:`gensim.corpora.wikicorpus.extract_pages` for the exact details
of the page context.
Parameters
----------
elem : etree.Element
XML etree element
text : str
The text of the XML node
namespace : str
XML namespace of the XML element
title : str
Page title
page_tag : str
XPath expression for page.
text_path : str
XPath expression for text.
title_path : str
XPath expression for title.
ns_path : str
XPath expression for namespace.
pageid_path : str
XPath expression for page id.
Example
-------
.. sourcecode:: pycon
>>> import gensim.corpora
>>> filter_func = gensim.corpora.wikicorpus.filter_example
>>> dewiki = gensim.corpora.WikiCorpus(
... './dewiki-20180520-pages-articles-multistream.xml.bz2',
... filter_articles=filter_func)
"""
# Filter German wikipedia dump for articles that are marked either as
# Lesenswert (featured) or Exzellent (excellent) by wikipedia editors.
# *********************
# regex is in the function call so that we do not pollute the wikicorpus
# namespace do not do this in production as this function is called for
# every element in the wiki dump
_regex_de_excellent = re.compile(r'.*\{\{(Exzellent.*?)\}\}[\s]*', flags=re.DOTALL)
_regex_de_featured = re.compile(r'.*\{\{(Lesenswert.*?)\}\}[\s]*', flags=re.DOTALL)
if text is None:
return False
if _regex_de_excellent.match(text) or _regex_de_featured.match(text):
return True
else:
return False
def find_interlinks(raw):
"""Find all interlinks to other articles in the dump.
Parameters
----------
raw : str
Unicode or utf-8 encoded string.
Returns
-------
list
List of tuples in format [(linked article, the actual text found), ...].
"""
filtered = filter_wiki(raw, promote_remaining=False, simplify_links=False)
interlinks_raw = re.findall(RE_P16, filtered)
interlinks = []
for parts in [i.split('|') for i in interlinks_raw]:
actual_title = parts[0]
try:
interlink_text = parts[1]
except IndexError:
interlink_text = actual_title
interlink_tuple = (actual_title, interlink_text)
interlinks.append(interlink_tuple)
legit_interlinks = [(i, j) for i, j in interlinks if '[' not in i and ']' not in i]
return legit_interlinks
def filter_wiki(raw, promote_remaining=True, simplify_links=True):
"""Filter out wiki markup from `raw`, leaving only text.
Parameters
----------
raw : str
Unicode or utf-8 encoded string.
promote_remaining : bool
Whether uncaught markup should be promoted to plain text.
simplify_links : bool
Whether links should be simplified keeping only their description text.
Returns
-------
str
`raw` without markup.
"""
# parsing of the wiki markup is not perfect, but sufficient for our purposes
# contributions to improving this code are welcome :)
text = utils.to_unicode(raw, 'utf8', errors='ignore')
text = utils.decode_htmlentities(text) # '&nbsp;' --> '\xa0'
return remove_markup(text, promote_remaining, simplify_links)
def remove_markup(text, promote_remaining=True, simplify_links=True):
"""Filter out wiki markup from `text`, leaving only text.
Parameters
----------
text : str
String containing markup.
promote_remaining : bool
Whether uncaught markup should be promoted to plain text.
simplify_links : bool
Whether links should be simplified keeping only their description text.
Returns
-------
str
`text` without markup.
"""
text = re.sub(RE_P2, '', text) # remove the last list (=languages)
# the wiki markup is recursive (markup inside markup etc)
# instead of writing a recursive grammar, here we deal with that by removing
# markup in a loop, starting with inner-most expressions and working outwards,
# for as long as something changes.
# text = remove_template(text) # Note
text = remove_file(text)
iters = 0
while True:
old, iters = text, iters + 1
text = re.sub(RE_P0, '', text) # remove comments
text = re.sub(RE_P1, '', text) # remove footnotes
text = re.sub(RE_P9, '', text) # remove outside links
text = re.sub(RE_P10, '', text) # remove math content
text = re.sub(RE_P11, '', text) # remove all remaining tags
# text = re.sub(RE_P14, '', text) # remove categories # Note
text = re.sub(RE_P5, '\\3', text) # remove urls, keep description
if simplify_links:
text = re.sub(RE_P6, '\\2', text) # simplify links, keep description only
# remove table markup
text = text.replace("!!", "\n|") # each table head cell on a separate line
text = text.replace("|-||", "\n|") # for cases where a cell is filled with '-'
text = re.sub(RE_P12, '\n', text) # remove formatting lines
text = text.replace('|||', '|\n|') # each table cell on a separate line(where |{{a|b}}||cell-content)
text = text.replace('||', '\n|') # each table cell on a separate line
text = re.sub(RE_P13, '\n', text) # leave only cell content
text = re.sub(RE_P17, '\n', text) # remove formatting lines
# remove empty mark-up
text = text.replace('[]', '')
# stop if nothing changed between two iterations or after a fixed number of iterations
if old == text or iters > 2:
break
if promote_remaining:
text = text.replace('[', '').replace(']', '') # promote all remaining markup to plain text
return text
def remove_template(s):
"""Remove template wikimedia markup.
Parameters
----------
s : str
String containing markup template.
Returns
-------
str
Сopy of `s` with all the `wikimedia markup template <http://meta.wikimedia.org/wiki/Help:Template>`_ removed.
Notes
-----
Since template can be nested, it is difficult remove them using regular expressions.
"""
# Find the start and end position of each template by finding the opening
# '{{' and closing '}}'
n_open, n_close = 0, 0
starts, ends = [], [-1]
in_template = False
prev_c = None
for i, c in enumerate(s):
if not in_template:
if c == '{' and c == prev_c:
starts.append(i - 1)
in_template = True
n_open = 1
if in_template:
if c == '{':
n_open += 1
elif c == '}':
n_close += 1
if n_open == n_close:
ends.append(i)
in_template = False
n_open, n_close = 0, 0
prev_c = c
# Remove all the templates
starts.append(None)
return ''.join(s[end + 1:start] for end, start in zip(ends, starts))
def remove_file(s):
"""Remove the 'File:' and 'Image:' markup, keeping the file caption.
Parameters
----------
s : str
String containing 'File:' and 'Image:' markup.
Returns
-------
str
Сopy of `s` with all the 'File:' and 'Image:' markup replaced by their `corresponding captions
<http://www.mediawiki.org/wiki/Help:Images>`_.
"""
# The regex RE_P15 match a File: or Image: markup
for match in re.finditer(RE_P15, s):
m = match.group(0)
caption = m[:-2].split('|')[-1]
s = s.replace(m, caption, 1)
return s
def tokenize(content):
# ORIGINAL VERSION
#def tokenize(content, token_min_len=TOKEN_MIN_LEN, token_max_len=TOKEN_MAX_LEN, lower=True):
"""Tokenize a piece of text from Wikipedia.
Set `token_min_len`, `token_max_len` as character length (not bytes!) thresholds for individual tokens.
Parameters
----------
content : str
String without markup (see :func:`~gensim.corpora.wikicorpus.filter_wiki`).
token_min_len : int
Minimal token length.
token_max_len : int
Maximal token length.
lower : bool
Convert `content` to lower case?
Returns
-------
list of str
List of tokens from `content`.
"""
# ORIGINAL VERSION
# TODO maybe ignore tokens with non-latin characters? (no chinese, arabic, russian etc.)
# return [
# utils.to_unicode(token) for token in utils.tokenize(content, lower=lower, errors='ignore')
# if token_min_len <= len(token) <= token_max_len and not token.startswith('_')
# ]
# NEW VERSION
return [token.encode('utf8') for token in utils.tokenize(content, lower=True, errors='ignore')
if len(token) <= 15 and not token.startswith('_')]
# TO RESTORE MOST PUNCTUATION
# return [token.encode('utf8') for token in content.split()
# if len(token) <= 15 and not token.startswith('_')]
def get_namespace(tag):
"""Get the namespace of tag.
Parameters
----------
tag : str
Namespace or tag.
Returns
-------
str
Matched namespace or tag.
"""
m = re.match("^{(.*?)}", tag)
namespace = m.group(1) if m else ""
if not namespace.startswith("http://www.mediawiki.org/xml/export-"):
raise ValueError("%s not recognized as MediaWiki dump namespace" % namespace)
return namespace
_get_namespace = get_namespace
def extract_pages(f, filter_namespaces=False, filter_articles=None):
"""Extract pages from a MediaWiki database dump.
Parameters
----------
f : file
File-like object.
filter_namespaces : list of str or bool
Namespaces that will be extracted.
Yields
------
tuple of (str or None, str, str)
Title, text and page id.
"""
elems = (elem for _, elem in iterparse(f, events=("end",)))
# We can't rely on the namespace for database dumps, since it's changed
# it every time a small modification to the format is made. So, determine
# those from the first element we find, which will be part of the metadata,
# and construct element paths.
elem = next(elems)
namespace = get_namespace(elem.tag)
ns_mapping = {"ns": namespace}
page_tag = "{%(ns)s}page" % ns_mapping
text_path = "./{%(ns)s}revision/{%(ns)s}text" % ns_mapping
title_path = "./{%(ns)s}title" % ns_mapping
ns_path = "./{%(ns)s}ns" % ns_mapping
pageid_path = "./{%(ns)s}id" % ns_mapping
for elem in elems:
if elem.tag == page_tag:
title = elem.find(title_path).text
text = elem.find(text_path).text
if filter_namespaces:
ns = elem.find(ns_path).text
if ns not in filter_namespaces:
text = None
if filter_articles is not None:
if not filter_articles(
elem, namespace=namespace, title=title,
text=text, page_tag=page_tag,
text_path=text_path, title_path=title_path,
ns_path=ns_path, pageid_path=pageid_path):
text = None
pageid = elem.find(pageid_path).text
yield title, text or "", pageid # empty page will yield None
# Prune the element tree, as per
# http://www.ibm.com/developerworks/xml/library/x-hiperfparse/
# except that we don't need to prune backlinks from the parent
# because we don't use LXML.
# We do this only for <page>s, since we need to inspect the
# ./revision/text element. The pages comprise the bulk of the
# file, so in practice we prune away enough.
elem.clear()
_extract_pages = extract_pages # for backward compatibility
def process_article(args):
# ORIGINAL VERSION
#def process_article(args, tokenizer_func=tokenize, token_min_len=TOKEN_MIN_LEN,
# token_max_len=TOKEN_MAX_LEN, lower=True):
"""Parse a Wikipedia article, extract all tokens.
Notes
-----
Set `tokenizer_func` (defaults is :func:`~gensim.corpora.wikicorpus.tokenize`) parameter for languages
like Japanese or Thai to perform better tokenization.
The `tokenizer_func` needs to take 4 parameters: (text: str, token_min_len: int, token_max_len: int, lower: bool).
Parameters
----------
args : (str, bool, str, int)
Article text, lemmatize flag (if True, :func:`~gensim.utils.lemmatize` will be used), article title,
page identificator.
tokenizer_func : function
Function for tokenization (defaults is :func:`~gensim.corpora.wikicorpus.tokenize`).
Needs to have interface:
tokenizer_func(text: str, token_min_len: int, token_max_len: int, lower: bool) -> list of str.
token_min_len : int
Minimal token length.
token_max_len : int
Maximal token length.
lower : bool
Convert article text to lower case?
Returns
-------
(list of str, str, int)
List of tokens from article, title and page id.
"""
text, lemmatize, title, pageid = args
text = filter_wiki(text)
if lemmatize:
result = utils.lemmatize(text)
else:
# ORIGINAL VERSION
# result = tokenizer_func(text, token_min_len, token_max_len, lower)
# NEW VERSION
result = tokenize(text)
# result = title + text
return result, title, pageid
def init_to_ignore_interrupt():
"""Enables interruption ignoring.
Warnings
--------
Should only be used when master is prepared to handle termination of
child processes.
"""
signal.signal(signal.SIGINT, signal.SIG_IGN)
def _process_article(args):
"""Same as :func:`~gensim.corpora.wikicorpus.process_article`, but with args in list format.
Parameters
----------
args : [(str, bool, str, int), (function, int, int, bool)]
First element - same as `args` from :func:`~gensim.corpora.wikicorpus.process_article`,
second element is tokenizer function, token minimal length, token maximal length, lowercase flag.
Returns
-------
(list of str, str, int)
List of tokens from article, title and page id.
Warnings
--------
Should not be called explicitly. Use :func:`~gensim.corpora.wikicorpus.process_article` instead.
"""
tokenizer_func, token_min_len, token_max_len, lower = args[-1]
args = args[:-1]
return process_article(
args, tokenizer_func=tokenizer_func, token_min_len=token_min_len,
token_max_len=token_max_len, lower=lower
)
class WikiCorpus(TextCorpus):
"""Treat a Wikipedia articles dump as a read-only, streamed, memory-efficient corpus.
Supported dump formats:
* <LANG>wiki-<YYYYMMDD>-pages-articles.xml.bz2
* <LANG>wiki-latest-pages-articles.xml.bz2
The documents are extracted on-the-fly, so that the whole (massive) dump can stay compressed on disk.
Notes
-----
Dumps for the English Wikipedia can be founded at https://dumps.wikimedia.org/enwiki/.
Attributes
----------
metadata : bool
Whether to write articles titles to serialized corpus.
Warnings
--------
"Multistream" archives are *not* supported in Python 2 due to `limitations in the core bz2 library
<https://docs.python.org/2/library/bz2.html#de-compression-of-files>`_.
Examples
--------
.. sourcecode:: pycon
>>> from gensim.test.utils import datapath, get_tmpfile
>>> from gensim.corpora import WikiCorpus, MmCorpus
>>>
>>> path_to_wiki_dump = datapath("enwiki-latest-pages-articles1.xml-p000000010p000030302-shortened.bz2")
>>> corpus_path = get_tmpfile("wiki-corpus.mm")
>>>
>>> wiki = WikiCorpus(path_to_wiki_dump) # create word->word_id mapping, ~8h on full wiki
>>> MmCorpus.serialize(corpus_path, wiki) # another 8h, creates a file in MatrixMarket format and mapping
"""
def __init__(self, fname, processes=None, lemmatize=utils.has_pattern(), dictionary=None,
filter_namespaces=('0',)):
# ORIGINAL VERSION
# filter_namespaces=('0',), tokenizer_func=tokenize, article_min_tokens=ARTICLE_MIN_WORDS,
# token_min_len=TOKEN_MIN_LEN, token_max_len=TOKEN_MAX_LEN, lower=True, filter_articles=None):
"""Initialize the corpus.
Unless a dictionary is provided, this scans the corpus once,
to determine its vocabulary.
Parameters
----------
fname : str
Path to the Wikipedia dump file.
processes : int, optional
Number of processes to run, defaults to `max(1, number of cpu - 1)`.
lemmatize : bool
Use lemmatization instead of simple regexp tokenization.
Defaults to `True` if you have the `pattern <https://github.com/clips/pattern>`_ package installed.
dictionary : :class:`~gensim.corpora.dictionary.Dictionary`, optional
Dictionary, if not provided, this scans the corpus once, to determine its vocabulary
**IMPORTANT: this needs a really long time**.
filter_namespaces : tuple of str, optional
Namespaces to consider.
tokenizer_func : function, optional
Function that will be used for tokenization. By default, use :func:`~gensim.corpora.wikicorpus.tokenize`.
If you inject your own tokenizer, it must conform to this interface:
`tokenizer_func(text: str, token_min_len: int, token_max_len: int, lower: bool) -> list of str`
article_min_tokens : int, optional
Minimum tokens in article. Article will be ignored if number of tokens is less.
token_min_len : int, optional
Minimal token length.
token_max_len : int, optional
Maximal token length.
lower : bool, optional
If True - convert all text to lower case.
filter_articles: callable or None, optional
If set, each XML article element will be passed to this callable before being processed. Only articles
where the callable returns an XML element are processed, returning None allows filtering out
some articles based on customised rules.
Warnings
--------
Unless a dictionary is provided, this scans the corpus once, to determine its vocabulary.
"""
self.fname = fname
self.filter_namespaces = filter_namespaces
# self.filter_articles = filter_articles
self.metadata = True
if processes is None:
processes = max(1, multiprocessing.cpu_count() - 1)
self.processes = processes
self.lemmatize = lemmatize
# self.tokenizer_func = tokenizer_func
# self.article_min_tokens = article_min_tokens
# self.token_min_len = token_min_len
# self.token_max_len = token_max_len
# self.lower = lower
# get_title = cur_title
if dictionary is None:
self.dictionary = Dictionary(self.get_texts())
else:
self.dictionary = dictionary
def get_texts(self):
"""Iterate over the dump, yielding a list of tokens for each article that passed
the length and namespace filtering.
Uses multiprocessing internally to parallelize the work and process the dump more quickly.
Notes
-----
This iterates over the **texts**. If you want vectors, just use the standard corpus interface
instead of this method:
Examples
--------
.. sourcecode:: pycon
>>> from gensim.test.utils import datapath
>>> from gensim.corpora import WikiCorpus
>>>
>>> path_to_wiki_dump = datapath("enwiki-latest-pages-articles1.xml-p000000010p000030302-shortened.bz2")
>>>
>>> for vec in WikiCorpus(path_to_wiki_dump):
... pass
Yields
------
list of str
If `metadata` is False, yield only list of token extracted from the article.
(list of str, (int, str))
List of tokens (extracted from the article), page id and article title otherwise.
"""
articles, articles_all = 0, 0
positions, positions_all = 0, 0
# ORIGINAL VERSION
# tokenization_params = (self.tokenizer_func, self.token_min_len, self.token_max_len, self.lower)
# texts = \
# ((text, self.lemmatize, title, pageid, tokenization_params)
# for title, text, pageid
# in extract_pages(bz2.BZ2File(self.fname), self.filter_namespaces, self.filter_articles))
# pool = multiprocessing.Pool(self.processes, init_to_ignore_interrupt)
# NEW VERSION
texts = ((text, self.lemmatize, title, pageid) for title, text, pageid
in extract_pages(bz2.BZ2File(self.fname), self.filter_namespaces))
pool = multiprocessing.Pool(self.processes)
try:
# process the corpus in smaller chunks of docs, because multiprocessing.Pool
# is dumb and would load the entire input into RAM at once...
# ORIGINAL VERSION
# for group in utils.chunkize(texts, chunksize=10 * self.processes, maxsize=1):
# NEW VERSION
for group in utils.chunkize_serial(texts, chunksize=10 * self.processes):
# ORIGINAL VERSION
# for tokens, title, pageid in pool.imap(_process_article, group):
# NEW VERSION
for tokens, title, pageid in pool.imap(process_article, group): # chunksize=10):
articles_all += 1
positions_all += len(tokens)
# article redirects and short stubs are pruned here
# ORIGINAL VERSION
# if len(tokens) < self.article_min_tokens or \
# any(title.startswith(ignore + ':') for ignore in IGNORED_NAMESPACES):
# NEW VERSION FOR ENTIRE BLOCK
if len(tokens) < ARTICLE_MIN_WORDS or \
any(title.startswith(ignore + ':') for ignore in IGNORED_NAMESPACES):
continue
articles += 1
positions += len(tokens)
try:
if self.metadata:
title = title.replace(' ', '_')
title = (title + ',')
title = bytes(title, 'utf-8')
tokens.insert(0,title)
yield tokens
else:
yield tokens
except Exception as e:
print('Wikicorpus exception error: ' + str(e))
except KeyboardInterrupt:
logger.warn(
"user terminated iteration over Wikipedia corpus after %i documents with %i positions "
"(total %i articles, %i positions before pruning articles shorter than %i words)",
# ORIGINAL VERSION
# articles, positions, articles_all, positions_all, self.article_min_tokens
# NEW VERSION
articles, positions, articles_all, positions_all
)
except PicklingError as exc:
raise_from(PicklingError('Can not send filtering function {} to multiprocessing, '
'make sure the function can be pickled.'.format(self.filter_articles)), exc)
else:
logger.info(
"finished iterating over Wikipedia corpus of %i documents with %i positions "
"(total %i articles, %i positions before pruning articles shorter than %i words)",
# ORIGINAL VERSION
# articles, positions, articles_all, positions_all, self.article_min_tokens
# NEW VERSION
articles, positions, articles_all, positions_all
)
self.length = articles # cache corpus length
finally:
pool.terminate()
Gensim provides well documented code that is written in an understandable way.
Most of the modifications I made occurred at the bottom of the code listing. However, the text routine at the top of the file allows us to tailor what page ‘sections’ are kept or not in each Wikipedia article. Because of their substantive lexical content, I add the page templates and category names to be retained with the text body.
Assuming I will want to retain these modifications and understand them at a later date, I block off all modified sections with ORIGINAL VERSION and NEW VERSION tags. One change was to remove punctuation. Another was to grab and capture the article title.
This file, then, becomes a replacement to the original wikicorpus.py code. I am cognizant that changing underlying source code for local purposes is generally considered to be a BAD idea. It very well may be so in this case. However, with the backups, and being attentive to version updates and keeping working code in sync, I guess I do not see where keeping track of a modification is any less sustainable than needing to update existing code to a modification. Both require inspection and effort. If I diff on the changed underlying module, I suspect it is of equivalent effort or lesser effort to change a third-party interface modification.
The net result is that I am now capturing the substantive content of these articles in a form I want to process.
Remove Stoplist
In my initial workflow, I had the step of stoplist removal later in the process because I thought it might be helpful to have all text prior to phrase identification. A stoplist (also known as ‘stop words‘), by the way, is a listing of very common words (mostly conjuctions, common verb tenses, articles and propositions) that can be removed from a block of text without adversely affecting its meaning or readability.
Since it proved superior to not retain these stop words when forming n-grams (see next section), I moved the routine up to be next processing of the Wikipedia pages. Here is the relevant code (NOTE: Neither the wikipedia-output-full.txt or wikipedia-output-full-stopped.txt files below are posted to GitHub due to their large (nearly GB) size; they can be reproduced using the GenSim standard stoplist as enhanced with the more_stops below):
import sys
from gensim.parsing.preprocessing import remove_stopwords # Key line for stoplist
from smart_open import smart_open
in_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-output-full.txt'
out_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-output-full-stopped.txt'
more_stops = ['b', 'c', 'category', 'com', 'd', 'f', 'formatnum', 'g', 'gave', 'gov', 'h',
'htm', 'html', 'http', 'https', 'id', 'isbn', 'j', 'k', 'l', 'loc', 'm', 'n',
'need', 'needed', 'org', 'p', 'properties', 'q', 'r', 's', 'took', 'url', 'use',
'v', 'w', 'www', 'y', 'z']
documents = smart_open(in_f, 'r', encoding='utf-8')
content = [doc.split(' ') for doc in documents]
with open(out_f, 'w', encoding='utf-8') as output:
i = 0
for line in content:
try:
line = ', '.join(line)
line = line.replace(', ', ' ')
line = remove_stopwords(line)
querywords = line.split()
resultwords = [word for word in querywords if word.lower() not in more_stops]
line = ' '.join(resultwords)
line = line + '\n'
output.write(line)
except Exception as e:
print ('Exception error: ' + str(e))
i = i + 1
if (i % 10000 == 0):
print('Stopwords applied to ' + str(i) + ' articles')
output.close()
print('Stopwords applied to ' + str(i) + ' articles;')
print('Processing complete!') Stopwords applied to 10000 articles
Stopwords applied to 20000 articles
Stopwords applied to 30000 articles
Stopwords applied to 31157 articles;
Processing complete!
Gensim comes with its own stoplist, to which I added a few of my own, including removal of the category keyword that arose from adding that grouping. The output of this routine is the next file in the pipeline, wikipedia-output-full-stopped.txt.
Phrase Identification and Extraction
Phrases are n-grams, generally composed of two or three paired words, which are known as ‘bigrams’ and ‘trigrams’, respectively. Phrases are one of the most powerful ways to capture domain or technical language, since these compounded terms arise through the use and consensus of their users. Some phrases help disambiguate specific entities or places, as when for example ‘river’, ‘state’, ‘university’ or ‘buckeyes’ does when combined with the term ‘ohio’.
Generally, most embeddings or corpora do not include n-grams in their initial preparation. But, for the reasons above, and experience of the usefulness of n-grams to text retrieval, we decided to include phrase identification and extraction as part of our preprocessing.
Again, gensim comes with a pre-trained phrase identifier (like all gensim models, you can re-train and tune these models as you gain experience and want them to perform differently). The main work of this routine is the ngram call, wherein term adjacency is used to construct paired term indentifications. Here is the code and settings for our first pass with this function to create our initial bigrams from the stopped input text:
import sys
from gensim.models.phrases import Phraser, Phrases
from gensim.parsing.preprocessing import remove_stopwords # Key line for stoplist
from smart_open import smart_open
in_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-output-full-stopped.txt'
out_f = r'C:\1-PythonProjects\kbpedia\v300\models\inputs\wikipedia-bigram.txt'
documents = smart_open(in_f, 'r', encoding='utf-8')
sentence_stream = [doc.split(' ') for doc in documents]
common_terms = ['aka']
ngram = Phrases(sentence_stream, min_count=3,threshold=10, max_vocab_size=80000000,
delimiter=b'_', common_terms=common_terms)
ngram = Phraser(ngram)
content = list(ngram[sentence_stream])
with open(out_f, 'w', encoding='utf-8') as output:
i = 0
for line in content:
try:
line = ', '.join(line)
line = line.replace(', ', ' ')
line = line.replace(' s ', '')
output.write(line)
except Exception as e:
print ('Exception error: ' + str(e))
i = i + 1
if (i % 10000 == 0):
print('ngrams calculated for ' + str(i) + ' articles')
output.close()
print('Calculated ngrams for ' + str(i) + ' articles;')
print('Processing complete!') ngrams calculated for 10000 articles
ngrams calculated for 20000 articles
ngrams calculated for 30000 articles
Calculated ngrams for 31157 articles;
Processing complete!
This routine takes about 14 minutes to run on my laptop, with the settings as shown. Note in the routine where we set the delimiter to be the underscore character; this is how we know the bigram.
Once this routine finishes, we can take its output and re-use it as input to a subsequent run. Now, we will be producing trigrams where we can match to existing bigrams. Generally, we set our thresholds and minimum counts higher. In our case, the new settings are min_count=8, threshold=50 The trigram analysis takes 19 min to run.
We have now completed our preprocessing steps for the embedding models we introduce in the next installment.
Additional Documentation
Here are many supplementary resources useful to the environment and natural language processing capabilities introduced in this installment.
- Transformers: State-of-the-art Natural Language Processing
- Corpus from a Wikipedia Dump
- Building a Wikipedia Text Corpus for Natural Language Processing.
PyTorch and pandas
- Convert Pandas Dataframe to PyTorch Tensor – pandas → numpy → torch
- PyTorch Dataset: Reading Data Using Pandas vs. NumPy
- See CWPK #56 for pandas/networkx reads and imports.
PyTorch Resources and Tutorials
- The Most Complete Guide to PyTorch for Data Scientists provides the basics of tensors and tensor operations and then provides a high-level overview of the PyTorch capabilities
- Awesome-Pytorch-list provides multiple resource categories, with 230 in related packages alone
- The official PyTorch documentation
- Building Efficient Custom Datasets in PyTorch
- Getting Started with PyTorch: A Deep Learning Tutorial
- Incredible PyTorch is a curated list of tutorials, papers, projects, communities and more relating to PyTorch
- Learning PyTorch with Examples.
spaCy and gensim
- Natural Language in Python using spaCy: An Introduction
- Gensim Tutorial – A Complete Beginners Guide is excellent and comprehensive.
*.ipynb file. It may take a bit of time for the interactive option to load.




Is the data closed? I am unsure where to look for wikipedia-output-full-stopped.txt for example
Hi Dean,
Yes, sorry, I did not note in the text that the files are too large to post to GitHub. (I have now updated the text.) You can reproduce the files (which are nearly 1 GB in size) using the standard Gensim stoplist plus the additions shown under ‘more_stop_’ in the code.